In conjunction with the National Holstein Convention, held at Pheasant Run Resort in St. Charles, Illinois, Holstein Association USA conducted a well-rounded panel on genomics that provided both technical insight and practical advice for Holstein breeders in attendance.
The panel consisted of four individuals and covered every facet of genomics – from the latest in gene expression to strategies for utilizing genomic data for on-farm management decisions.
Although genomics entered the industry five years ago, the panel drew a crowd interested and ready to hear more as genomic technology develops and confidence continues to grow. In introducing the topic, John Meyer, CEO of Holstein Association USA, provided evidence of the increased uptake by Holstein breeders with the staggering number of genomic tests conducted in 2014.
According to the USDA and the Council for Dairy Cattle Breeding (CDCB), there were 198,885 genomic tests performed on Holstein females and 30,283 Holstein males – equating to 86 percent of all genomic tests run on dairy cattle.
And while each of the four panelists presented separately to provide insight into their day-to-day interaction with genomics, all spoke in unison about the bright future and important role genomics will have in making a more profitable Holstein cow.
The panelists included Dr. Michael Bishop, senior market development specialist at Illumina; Chuck Ripp, owner and partner of Ripp’s Dairy Valley LLC in Dane, Wisconsin; Dr. Dan Weigel, director of outcomes research at Zoetis; and Dr. Nate Zwald, U.S. general manager for Alta Genetics.

A geneticist: Unravelingthe genome for future generations
Dr. Michael Bishop, senior market development specialist at Illumina
Illumina has been involved in bovine genomics from the start as the DNA sequencing company worked closely with the USDA, University of Missouri and University of Alberta in the development of the bovine 50k SNP chip. Today, Illumina-made chips are delivered to the dairy industry through its partners, like Zoetis.
Bishop explained that while we’re just beginning to understand about how genes are associated and expressed, there are projects in the pipeline that will begin to lead the industry into important discussions and decisions.
Currently in progress, the 1,000-bull genome project is a database of fully sequenced bovines that will serve as a pivot point for comparison and will help us understand and identify important genes and identification of causal mutations responsible for trait variation.
In time, the resulting progress and discovery made will give us the opportunity to go further, Bishop said, and realize the potential of personalized diagnostics and even gene editing. He posed a relevant example of gene editing that could benefit the entire industry: Could a simple base-pair change result in naturally hornless (or polled) cattle?
While it remains to be seen what will arise, Bishop reminded attendees that with this incredibly powerful tool, it’s important to remain accountable to our decisions impacting future generations.
“Successful animal production results from the combined application of art, science and good animal husbandry skills achieved from knowledge, hard work, experience and ... common sense,” he said.

A Holstein breeder: Making progress with a whole-herd approach
Chuck Ripp, owner and partner of Ripp’s Dairy Valley LLCin Dane, Wisconsin
Located in Dane, Wisconsin, Ripp’s Dairy Valley LLC has a herd of 1,000 cows and 900 heifers. Their rolling herd average is 31,100 pounds of milk, with 3.8 percent fat and 3 percent protein, and with a somatic cell count average of 170,000.
Ripp explained that, early on, he recognized advantages of genomics for their dairy, and they began heavily testing the herd. In fact, of the 550 heifer calves born on the farm each year, he estimates all but 5 percent are genomic-tested.
The farm’s mission is to do the best they can to milk the best 1,000 cows at Ripp’s Dairy Valley, and utilizing genomics supports this, according to Ripp. He explained, “For us, it’s not just about finding the high ones, but using genomic information to improve the herd as a whole.” In addition to confirming the sire and dam to ensure each animal is correctly identified, genomic testing aids Ripp in making management decisions.
He implements breeding protocols after receiving genomic test results – with the top 5 percent being flushed, next 25 percent being bred to sexed semen, lower 40 percent bred to conventional semen, and the bottom 30 percent carrying eggs. Ripp feels this allows him to multiply their top animals to advance the herd by improving production and profitability down the line.
While Ripp is not currently culling any heifers, he looks forward to utilizing genomic data to make more informed culling decisions. He emphasized that he is continuing to learn more about genomics, as he is certain genomic information will continue to be important.

A genotyping solutions provider: Analyzing and applying datafor real-world progress
Dr. Dan Weigel, director of outcomes research at Zoetis
Weigel began his presentation by reviewing the CDCB genetic reference base published with the December 2014 base change, which allows us to compare breeds. As the dominant breed, the numbers shown in Table 1 are the deviation from the average Holstein cow.
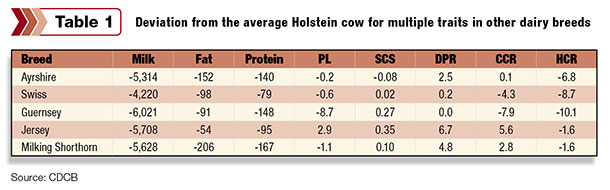
Weigel commended the Holstein breed on its strong advantage in milk, fat and protein, and also reviewed areas of opportunity for Holsteins in productive life (PL), cow conception rate (CCR) and heifer conception rate (HCR).
While these three traits are identified as lowly heritable traits, Weigel argues genomics can help make genetic progress. While HCR has a heritability of only 0.01, Figure 1 shows the results of a farm that genomic-tested 200 heifers.
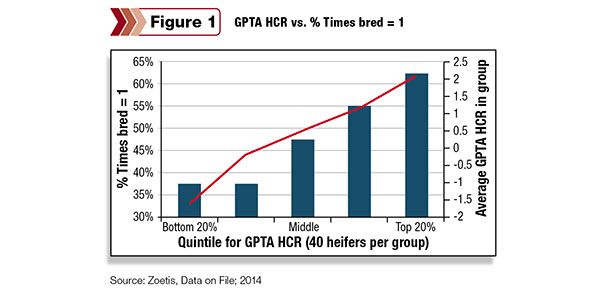
There’s a strong correlation between the gPTA HCR value and the number of times bred – providing breeders with confidence that genomics can reliably identify the lower end, even for lowly heritable traits.
Weigel emphasized that it’s not just about possessing the additional genetic information provided by genomic testing but also having a plan on how the information will inform your strategy and translate to the day-to-day decisions regarding selection, management and mating to help make genetic progress in the herd.
And, true to the Holstein USA magnet on Weigel’s fridge, he said that genomics reaffirms there are genetic differences among cows and “better cows make more money.”

An A.I. company: Striving to continue the global standard
Dr. Nate Zwald, U.S. general manager for Alta Genetics
Zwald began his comments by recognizing the U.S. Holstein breed as the envy of the world. In his work-related travels to 15 countries, Zwald was always asked about what was new in the U.S. in management and genetics.
Because of the robust infrastructure and powerful knowledge base of breeders, allied industry and even the youth, excitement of what’s next globally oftentimes originates with the U.S. He said, “Be proud of yourselves for that, but also recognize, we have to work hard to keep that.”
The U.S. Holstein has not always been the standard for the rest of the world. Zwald reminded breeders that the U.S. once imported Friesians at a time when they were the standard. While Europe kept the breed dual-purpose, the U.S. worked to make progress with production. He explained, “We made a more profitable cow than the Friesian. Genetically, we selected for the key traits that made a difference and made more profit.”
However, Zwald challenged, history tends to repeat itself, so what could happen to allow someone to catch us? There are opportunities for other breeds and other countries to catch up if we don’t maintain our focus on overall profitability. Zwald believes there’s excitement surrounding genomic technology, and by aligning overall on the ideal Holstein cow, every individual can apply it to their specific breeding goals. PD
Sarah Lenkaitis is a freelance writer and dairy farmer in Saint Charles, Illinois.




